DNA is the genetic code inside each of our cells, which we inherit from our parents.
The DNA in each of our cells is identical, yet different cells in our body have very distinct functions: skin cells have different functions from those of liver cells, heart muscle cells, or breast tissue cells. Therefore, how can the function and morphology of the cells be different when they all share the same code?
The answer is epigenetics, which is an extra layer of information superimposed on the DNA. Specifically, we look at DNA methylation patterns. Like DNA, we also inherit epigenetic information from our parents, which can be modified throughout life. This happens, for example, during embryonic development, when different cell types of our bodies start to emerge: different cells have different epigenetic signatures (i.e., epigenetic patterns are cell type-specific). While DNA can be considered as the “hardware” of a computer, DNA methylation is comparable to the software that defines which programs are run.
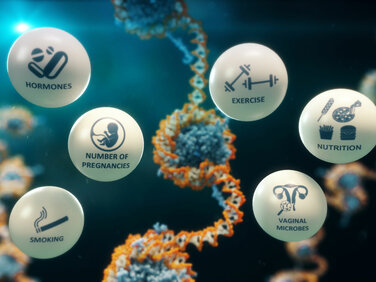
We now know that DNA methylation is not only modified during development but can be changed through a variety of other factors, including ageing, nutrition, exercise, socio-economic status, obesity, smoking, and exposure to hormones and chemicals (even while we are in the womb). Our work has identified signatures associated with environmental exposures.
As DNA methylation integrates genetic information (it is inherited) and non-genetic information (it can be modified by external factors), it is a highly promising candidate for a risk prediction tool, and importantly, cancer cells have distinct methylation patterns which can exist years before diagnosis. We can therefore use the epigenome to monitor the presence of cancer, and recent work suggests that the epigenome may even be able to predict the risk for the future development of cancers. This can be carried out using the “target tissue” of disease itself (e.g., breast tissue for breast cancer), but this is invasive and therefore not feasible for routine population-based screening. Recent work suggests that we may be able to use “surrogate” tissues, such as epithelial cells derived from buccal swabs, to predict individual cancer risk.
In several recent research papers, we have demonstrated the value of these DNA methylation signatures for women’s cancer risk prediction. We are currently working to validate these tools for clinical screening and identify whether we can also use them to monitor risk over time.